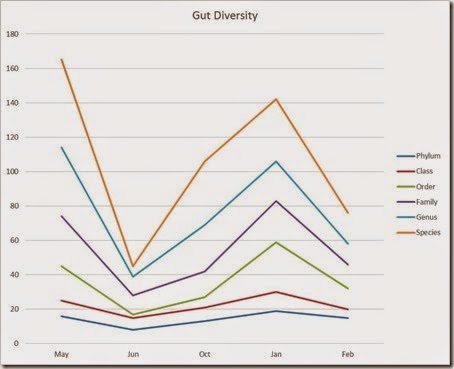
My gut diversity through time
Clark Ellis posts a nice summary of his uBiome results over at the uBiome Blog and now, with more detail at The Self-Taught Author blog. A long period of antibiotic use has made him acutely interested in the understanding gut diversity, so he asks others to post their uBiome diversity results too.
Here’s mine:
A few caveats:
- These values only represent the identified results, which generally bounce from about 70% (at the genus level) to 95% (phylum). There could well be dozens, perhaps thousands, of other unique bacteria that are simply too rare to be counted by the uBiome technology.
- A single bacterium can have a big effect, so it probably doesn’t mean much to look at raw counts. Remember that the mammalian genus canis includes wolves, coyotes, and jackals in addition to your trusty dog Fido. Simply knowing there’s a canis at the door tells you nothing about whether it’s safe to go out.
- Species information is (probably) meaningless. uBiome uses 16S rRNA technology that can’t differentiate below the genus level. They don’t even post species information on their web viewer; you have to uncover it from the raw data like I did. They claim it’s “experimental”, which I interrupt to mean they apply some statistical “guess”, perhaps based on general trends. Anyway, you shouldn’t rely on it.
Something strange happened in my June sample, which was taken three weeks after the one from May, in what was frankly a boring period of my life (no travel, no unusual food, no camping, etc.). It’s possible that result was simply a mistake.
Note: all of my data is posted on GitHub, and you’re welcome to explore it and compare to your heart’s content as long as you promise to let me know if you find anything interesting!